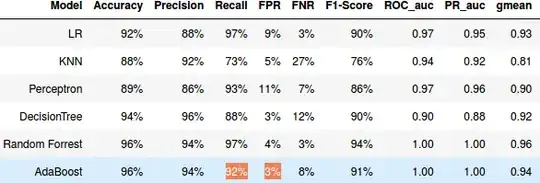
I am working on binary classification problem, I try to evaluate the performance of some classification algorithms (LR,Decission Tree , Random forest ...). I am using a cross validation technique (to avoid over-fitting) with AUC ROC as scoring function to compare the performance of the algorithms, but I am getting a weird results with Random forest and AdbBoost, I have a perfect AUC_ROC score (i.e. =1) despite the fact that the recall(TPR) and FPR of this algorithms are different from 1 and 0 respectively .
def FPR(y_true, y_pred):
tn, fp, fn, tp = confusion_matrix(y_true, y_pred).ravel()
result = fp / (fp+tn)
return result
def FNR(y_true, y_pred):
tn, fp, fn, tp = confusion_matrix(y_true, y_pred).ravel()
result = fn / (tp+fn)
return result
FPR_scorer = make_scorer(FPR)
FNR_scorer = make_scorer(FNR)
def get_CrossValResults2(model,cv_rst,bestIndx):
best=pd.DataFrame.from_dict(cv_rst).iloc[bestIndx]
roc="{:.12f}".format(best['mean_test_roc_auc'])
acc ="{:.0%}".format(best['mean_test_accuracy'])
prec ="{:.0%}".format(best['mean_test_precision'])
rec ="{:.0%}".format( best['mean_test_recall'])
f1 ="{:.0%}".format(best['mean_test_f1'])
r2="{:.2f}".format(best['mean_test_r2'])
g_mean="{:.2f}".format(best['mean_test_gmean'])
pr_auc="{:.8f}".format(best['mean_test_pr'])
fnr="{:.0%}".format(best['mean_test_fnr'])
fpr="{:.0%}".format(best['mean_test_fpr'])
rst = pd.DataFrame([[ model, acc,prec,rec,fpr,fnr,f1,roc,pr_auc,g_mean,r2]],columns = ['Model', 'Accuracy', 'Precision', 'Recall','FPR','FNR', 'F1-Score','ROC_auc','PR_auc','gmean','r2'])
return rst
cross_val_rst = pd.DataFrame(columns = ['Model', 'Accuracy', 'Precision', 'Recall','FPR','FNR', 'F1-Score','ROC_auc','PR_auc','gmean','r2'])
scoring = {'accuracy':'accuracy','recall':'recall','precision':'precision','fpr':FPR_scorer,'fnr':FNR_scorer,'f1':'f1' ,'roc_auc':'roc_auc','pr':'average_precision','gmean':Gmean_scorer,'r2':'r2'}
param_grid = {'n_estimators': [200],
'max_depth': [80,90],
'min_samples_leaf': [2,3, 4],
'min_samples_split': [2,5,12],
'criterion': [ 'gini'],
'class_weight' : [class_weights], 'n_jobs' : [-1]}
clf = GridSearchCV(RandomForestClassifier(class_weight=class_weights), param_grid, cv=kfold,scoring=scoring,refit=refit)#Fit the model
bestmodel = clf.fit(X,Y)
cross_val_rst = cross_val_rst.append(get_CrossValResults2(model='Random Forrest',bestIndx=bestmodel.best_index_,cv_rst=bestmodel.cv_results_),ignore_index=True)