I am currently trying to make a model to classify brain tumor patients by incidence of epilepsy using a combination of variables extracted from clinical records, and radiomics features from segmented MR images. In total there are 1396 features obtained from 53 patients, which have been min max scaled and z-score normalized (there were some categorical variables which have been one hot encoded). Based on existing literature in the field, I opted to use a logistic regression with an elastic net regularization to select the important features.
First, I tried to find the optimal alpha and L1 ratio from a set of values, using stratified kfold validation (there were more negatives than positives):
skf = StratifiedKFold(n_splits=5)
def scores_cv(model):
scores = cross_val_score(model, X, y, scoring='roc_auc', cv = skf)
return(scores)
alphas = [0.001, 0.005, 0.001 ,0.005, 0.009, 0.01, 0.1, 0.5, 1] #
l1_ratios = [1, 0.97 ,0.95, 0.93, 0.9, 0.5, 0.1, 0.01]
cv_elastic = [scores_cv(SGDClassifier(loss="log_loss", penalty="elasticnet",alpha = alpha,l1_ratio = l1_ratio)).mean()
for (alpha, l1_ratio) in product(alphas, l1_ratios)]
matplotlib.rcParams['figure.figsize'] = (12.0, 6.0)
idx = list(product(alphas, l1_ratios))
p_cv_elastic = pd.Series(cv_elastic, index = idx)
p_cv_elastic.plot(title = "Validation - Just Do It")
plt.xlabel("alpha - l1_ratio")
plt.ylabel("roc_auc")
print('alpha, lambda and error value, respectively = '+ str(p_cv_elastic[p_cv_elastic == p_cv_elastic.max()]))
Then, I fit a logistic regression model with the best hyperparameters on all the data, and looked for non-zero coefficients in the model.
enetlog = SGDClassifier(loss="log_loss", penalty="elasticnet",alpha = 0.1,l1_ratio = 0.93).fit(X,y)
print('accuracy:' +str(enetlog.score(X,y)))
y_pred = enetlog.predict(X)
print('stratified kfold accuracies:'+ str(scores_cv(enetlog)))
cm = confusion_matrix(y, y_pred, labels=enetlog.classes_)
disp = ConfusionMatrixDisplay(confusion_matrix=cm,
display_labels=enetlog.classes_)
disp.plot()
plt.show()
enet_results = pd.DataFrame(list(zip(list(X.columns),list(np.transpose(enetlog.coef_)))),columns=['Variable Name','Linear Regression Coefficient']) #if pipeline not used, it should be regr.coef_
enet_results = enet_results.sort_values(by=['Linear Regression Coefficient'])
enet_results = enet_results.loc[~((enet_results['Linear Regression Coefficient'] == 0) )]
enet_results['Linear Regression Coefficient'] = enet_results['Linear Regression Coefficient'].astype(float)
ax = enet_results.plot.barh(x='Variable Name', y='Linear Regression Coefficient', rot=0,figsize=(10, 15),fontsize = 12)
ax.bar_label(ax.containers[0],label_type='center')
The problem is most of the time I run the loop to find the best model, I end up with a different set of hyperparameters/maximum score. For instance the graph of metric against hyperparameter pair can look like this:

Then in another run it would look like this:
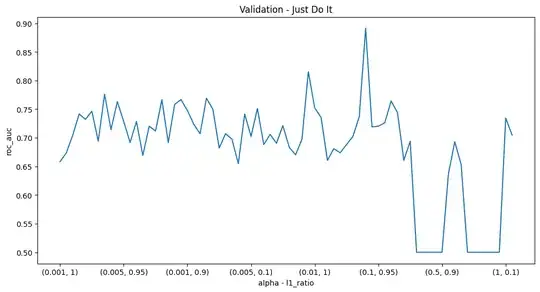 The best alpha, l1 ratio pair will usually be around 0.1,0.9 or 0.1,1. Then, when I pick the set of hyperparameters to construct the logistic regression model, there is often a different set of variables with nonzero coefficients in the model. Sometimes the model will have as low as 8 nonzero coefficients, and other times as high as 40.
Changing max_iter in either step (I tried up to max_iter = 10 000 000)does not seem to solve the inconsistencies between runs. I suspect the problem is a lack of data (only 53 samples), but I'm not sure if there are any other causes.
The best alpha, l1 ratio pair will usually be around 0.1,0.9 or 0.1,1. Then, when I pick the set of hyperparameters to construct the logistic regression model, there is often a different set of variables with nonzero coefficients in the model. Sometimes the model will have as low as 8 nonzero coefficients, and other times as high as 40.
Changing max_iter in either step (I tried up to max_iter = 10 000 000)does not seem to solve the inconsistencies between runs. I suspect the problem is a lack of data (only 53 samples), but I'm not sure if there are any other causes.
Thanks for your help.