Applying dimensional reduction could help understand why the neural network has made classifications, but we have to deep dive into the results using analytics.
In this example from Restnet50, you can see the UMAP result at each step. If you take the flattened one, you can see some outliers in different clusters. You would have something similar for the Melona classification.
The advantage of using UMAP is to detect why some classifications are wrong and why they are far from their correct cluster.
Therefore, you can get the ranges of each cluster (min and max values) and compare them with the wrong classified results.
For instance, if there is a wrongly classified melanoma in a cluster and you find out that the age was very different from the other cluster's age, you might emphasize the "age weight" with a power factor.
Unfortunately, it could be quite complex to do because all features are inter-dependants, and modifying one feature could have unforeseeable impacts on the others.
That's why many researchers try many potential solutions and choose the best one, without having a rigorous scientific approach because of the noise in the human-made data and the important quantity of learning iterations + the use of different modules.
In conclusion, applying dim reduction could give you a better understanding of a model and find new ideas to improve it thanks to the clusters' organization (overall in UMAP, where there is meaningful logic between clusters places), but such improvement could be very complex and could require many trials and fails.
Here are interesting articles about NN interpretability:
https://www.kaggle.com/code/subinium/interpretable-cnn-with-shap-mnist
https://towardsdatascience.com/dense-or-convolutional-part-2-interpretability-c310a9fd99a5
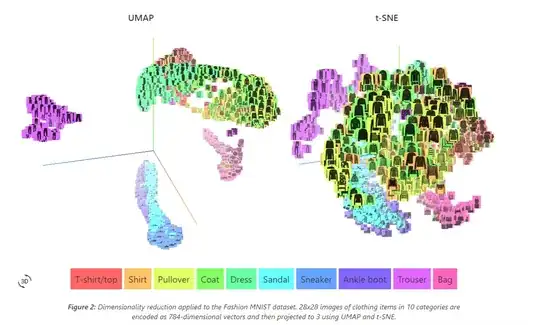
In this picture, you can see that UMAP has meaningful placement between clusters.