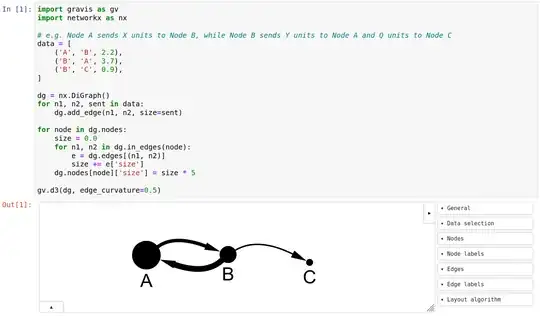
this is a more general question asking about Python libraries that are able to showcase networks with following criterias:
- Different size of the nodes depending on a metric (e.g. sum of some variable X received by all other nodes)
- Direction of the connection between the nodes (e.g. Node A sends X units to Node B, while Node B sends Y units to Node A and Q units to Node C) - So a node can receive and send at the same time.
- The connectors may vary in "thickness" relative to each other depending how many units they receive/send
- Dynamic UI - Network should be dynamic for e.g. filters like date ranges etc. that can be selected by the user.
I checked for Networkx and Plotlty, however they do not satisfy the direction criteria of the connectors.
Someone any idea?
A representation like this would work as well: https://viz.ged-project.de/