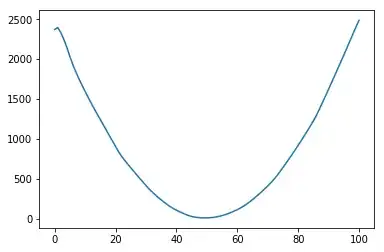
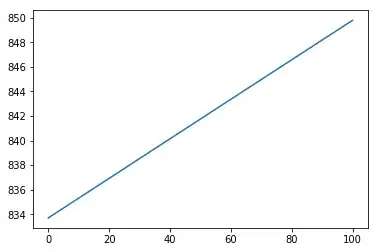
I noticed that feature scaling can destroy completely neural networks performance in some cases. Below are my results that you can reproduce easily.
I use a neural network to approximate function $g$ on $[0,100]$ as follows:
$\mathcal{N}\mathcal{N}(X; \theta) \approx g(X)$
where $X \sim \mathcal{U}([0,100])$. Then I randomly sample some $X$ ($(X_i)_i$) to be my training set, and use $Y_i=g(X_i)$ as targets.
Typically $\mathcal{N}\mathcal{N}$ is a Keras Sequential model, and has a few hidden layers with 15-20 neurons (elu), and a single output layer with no activation function. Parameters are initialized using kernel_initializer='normal'.
Now I noticed the following while testing my approximations in 1dim (before trying to generalize to higher dimensions):
For $g(x)=\max(0, X-50)$, scaling $X$ by dividing it by 100 does not really change the performance of the $\mathcal{N}\mathcal{N}$, which estimates $g$ relatively well.
However, when I use other functions such as $g(x)=1-\mathbb{I}_{x\in[40,60]}$ or $g(x)=(x-50)^2$, it fails completely.
Do you have an explanation?
This is very disturbing because how can you trust your network to approximate high-dimensional functions if it fails completely for simple 1-dimensional examples?
Below is an example implementation in Python:
from keras.models import Sequential
from keras.layers import Dense
import numpy as np
import matplotlib.pyplot as plt
d=1
nn_value_batch_size=128
nn_value_epochs=50
nn_value_training_size=10000
def nn_value_get_architecture(layers_sizes):
nn_value = Sequential()
nn_value.add(Dense(layers_sizes[0], input_dim=d, #d=1
kernel_initializer='normal', activation='elu'))
# hidden layers
for nb in layers_sizes[1:]:
nn_value.add(Dense(nb, kernel_initializer='normal',
activation='elu'))
# output layer
nn_value.add(Dense(1, kernel_initializer='normal'))
nn_value.compile(loss="mean_squared_error", optimizer='adam')
return nn_value
def nn_value_optimize(nn_value, objective_function,
x_normalize=True):
# fit the model
x_distribution = lambda size: np.random.uniform(0, 100, size)
for epoch in range(nn_value_epochs):
X_train = x_distribution(nn_value_training_size)
y_train = objective_function(X_train)
if x_normalize==True:
X_train = X_train/100
nn_value.fit(X_train, y_train,
batch_size=nn_value_batch_size,
verbose=1)
return nn_value
def g(X):
#return 1-(np.maximum(0., X-40)/(X-40) - np.maximum(0., X-60)/(X-60))
return (X-50)**2
#return np.maximum(0., X-50) #only example that works
nn_value = nn_value_get_architecture(
layers_sizes=[20,15,15,15,15])
g = np.vectorize(g)
normalize=True
nn_value = nn_value_optimize(nn_value, g, normalize)
# plot results
x = np.arange(0,101,1)
if normalize==True:
plt.plot(x, nn_value.predict(x/100))
else:
plt.plot(x, nn_value.predict(x))
plt.show()
No scaling:
With scaling: